Welcome to PoseBench’s documentation!¶




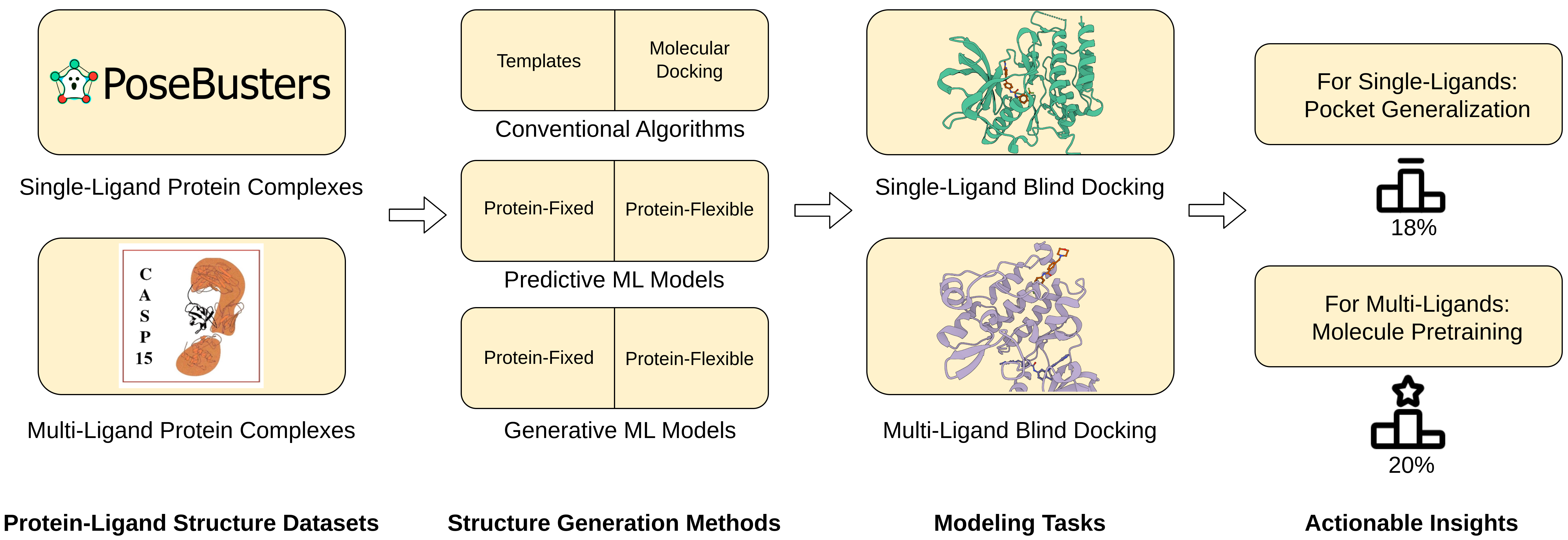
Comprehensive benchmarking of protein-ligand structure prediction methods
Contents:
- Installation
- Tutorials
- How to prepare PoseBench data
- Downloading Astex, PoseBusters, DockGen, and CASP15 data
- Downloading benchmark method predictions
- Downloading benchmark method interactions
- Downloading sequence databases (required only for RoseTTAFold-All-Atom inference)
- Downloading PDB metadata
- Predicting apo protein structures using ESMFold (optional, preprocessed data available)
- Available inference methods
- How to run a sweep of benchmarking experiments
- How to run inference with individual methods
- How to run inference with
DiffDock - How to run inference with
FABind - How to run inference with
DynamicBind - How to run inference with
NeuralPLexer - How to run inference with
FlowDock - How to run inference with
RoseTTAFold-All-Atom - How to run inference with
Chai-1 - How to run inference with
Boltz - How to run inference with
AlphaFold 3 - How to run inference with
AutoDock Vina - How to run inference with
TULIP
- How to run inference with
- How to run inference with a method ensemble
- How to create comparative plots of inference results
- For developers
- Acknowledgements
- Citing this work
- Bonus

